This article offers an introduction to nutritional genomics. Are you a nutritional or health professional working hard to recommend individualised dietary protocols for each of your clients? Maybe you have tried a food regime or which worked wonders for one of your loved ones – mother, aunt, sister – but did not achieve the same results for the other members of your family? Consider this, does everyone in your family wear the same shoe size? The problem is – one size may not fit all – this rule applies to both shoes and our nutrition.
Let’s start our introduction to nutritional genomics… from the very beginning!
The correct state of nutrition provides our body with optimal conditions for everyday functioning and helps maintain our health. It is known that a deficiency in some nutrients increases the risk of diseases. We can reduce this risk by increasing the intake of specific nutrients, ultimately reaching a level that will be beneficial. However, it is necessary to bear in mind that for many nutrients too high consumption is no longer profitable and starts to be harmful, leading again to the increased risk of disease. It is therefore essential to consider optimum levels of nutrient intake which are safe but also effective and supportive to optimum wellness.
These are used to design the UK’s food-based guidelines such as the Eatwell guide and food labelling to help people make healthy dietary choices. The DRVs are not intended for individuals with specific health needs. Most recommendations are classified according to gender and age, but these are not the only factors that should be considered when advising on diet and lifestyle. If the majority of our nutritional healthcare is currently based on what happens to the average person then how does this relate to you as an individual?
Through scientific research it has become clearer that the response to a particular nutrient is not the same for each person. There are a variety of reasons for this such as metabolic, environmental and/or genetic.
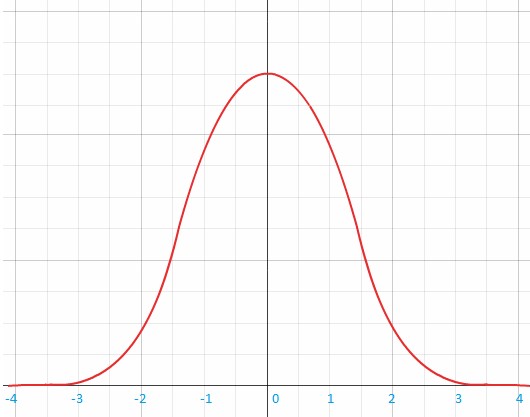
Why is it that people react differently to foods and certain nutrients? The benefit-risk response of nutrient intake was illustrated by John E. Hesketh et al. in figure 2. It shows that for a given nutrient there is a range of intake that provides a ‘window of benefit’, which varies between people.
Nutritional genomics
Precision Nutrition is a relatively new term that can be considered as a way to use information we continue to learn about our genomics in order to customise nutritional advice. Progress in scientific and medical research has led to understanding our biology as a whole system, known as the -omics era. The knowledge we can apply from -omics technology, together with understanding of nutrient-gene interactions has developed a field known as Nutritional Genomics.
At present, the nutritional genomic relationship is made up of two related but distinct fields namely, Nutrigenomics and Nutrigenetics. Nutrigenomics focuses on how diet affects gene expression. Nutrigenetics focuses on how the gene variants in our DNA influences our response to nutients i.e. how our genes determine the effects what our food has on us. Both aim to elucidate how the genome interacts with nutrition to influence genotype to optimise health through the application of personalised and precision nutrition.
Nutritional Genomics hold much promise for providing better nutritional advice to individuals. It does however require a deep understanding of nutrition, genetics, biochemistry and new ‘omic’ technologies and their relevance to the practice of preventive approaches for optimising health and preventing or delaying onset of disease. Before getting into the topic of nutritional genomics, let’s do a quick review of key terms.
DNA (Deoxyribonucleic acid) is a molecule, that constitutes our genetic template. DNA stores information and instructions for all organisms to grow, develop, function, and reproduce. You probably remember its construction from biology classes, but a small reminder never hurts!
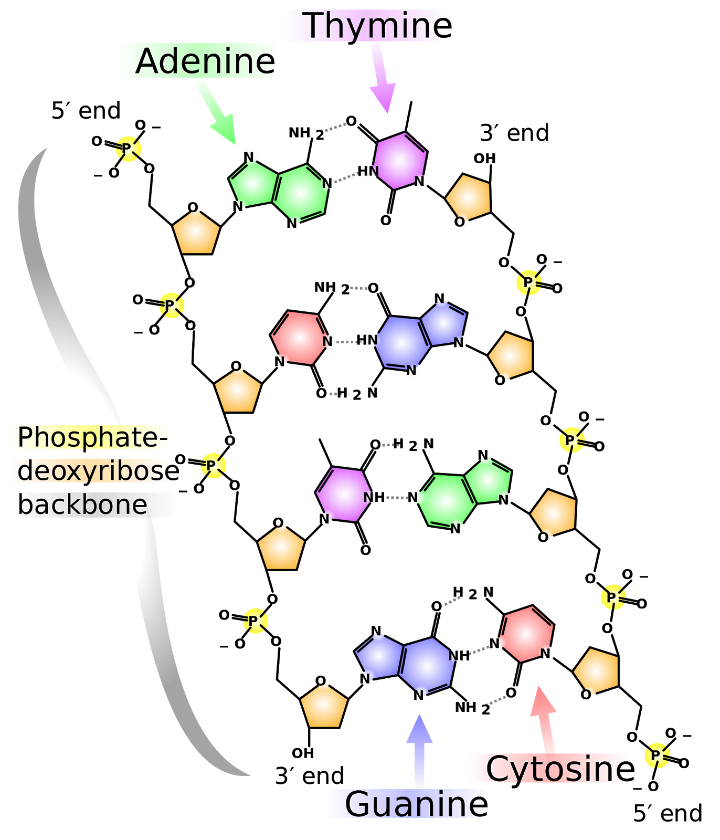
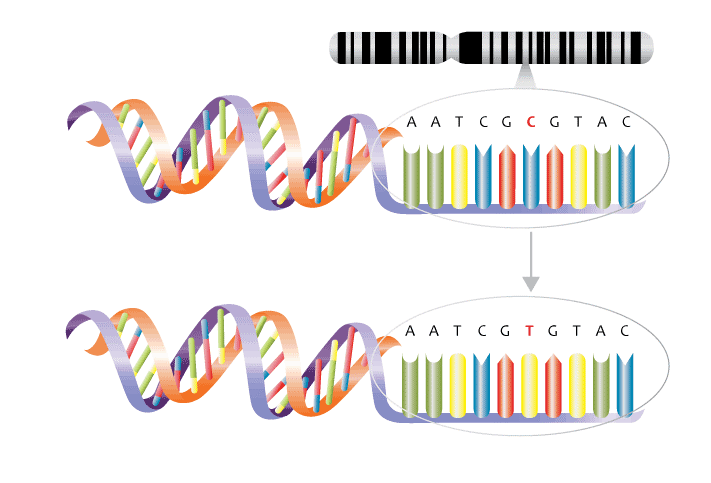
As shown in figure 3 below, DNA is composed of four different types of nucleotides bases – two double-ringed purine bases adenine (A) and guanine (G) and two single ringed pyrimidine bases, cytosine (C) and thymine (T).
Nucleotides are linked together by covalent phosphodiester bonds that link phosphate and deoxyribose’s to form a backbone chain. The four different bases are attached to this chain.
Complementary base pairing occurs between A and T (two hydrogen bonds) and between G and C (three hydrogen bonds) to form a stable double-stranded polymer.
Genes are regions of DNA that has the information essential for making specific proteins. Names of genes are written in italics, which allows us to distinguish them from the names of their corresponding proteins.
Giving an example will be much easier to understand:
- The TCF7L2 gene (written in italics) codes for a protein called TCF7L2 (transcription factor 7-like 2)
Gene expression is the process of converting information encoded in DNA into proteins (or other products in the form of various RNA structures). The production of proteins is divided into 2 steps, which are explained by the central dogma of molecular biology:
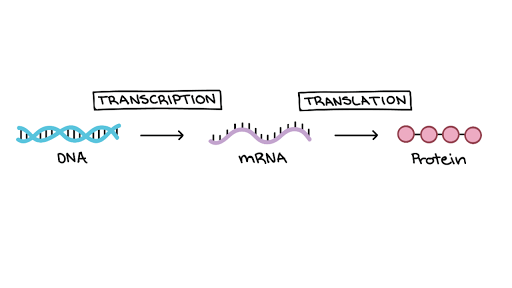
- Transcription – making RNA, rewriting the information contained in DNA. At this stage, DNA is separated into 2 complementary RNA strands, like a zipper, by enzymes called polymerases.
- Translation – protein synthesis based on the instructions from RNA.
A genome is the complete genetic information of an organism. To give you some historical perspective on this and to show how fast science and technologies are developing:
In 1990, The Human Genome Project was established as a worldwide scientific project to map the order of every nucleotide in the human genome. By 2000, President Bill Clinton and the British Prime Minister Tony Blair, announced that a ‘rough draft’ of the human genome had been completed and was declared successfully completed in 2003. It had taken 13 years to complete a complete genetic blueprint for one human being. The human genome was found to be approximately 3 billion nucleotides, around 22 000 genes, a large amount of information! Technology has rapidly evolved and in 2007, commercial genetic testing services – such as 23andMe – became publicly available. This was the breakthrough moment where genetic testing and individualised counseling became possible and the market has grown rapidly.
Nowadays, it is comparatively cheap, fast and easy to get your genome examined as modern testing methods do not need to check the whole genome. A common genetic testing, method involves checking known genetic variants or single nucleotide polymorphism (SNP). SNPs are the most common type of genetic variation among people and are a swap in the DNA code of one nucleotide for another.
Over 20 million validated SNPs have been identified, so how do we understand which ones are relevant to nutrigenetics?
Different experimental approaches can be used to identify genetic variants that modify the effects of dietary factors or influence food preferences. Using a candidate gene approach is a common method whereby a gene is selected based on its known or putative function. Depending on the number of SNPs in the gene, and whether any of them have known functional effects, analyses can be conducted using individual SNPs or combinations of SNPs. This could be considered a bottom-up approach which relies on researchers having a hypothesis of likely nutrigenetic associations. Recent studies have begun to apply a top-down approach known as genome-wide association studies (GWAS) to identify previously unknown genetic variants that could modify the response to diet. Both methods compare SNP data from groups of controls compared to patients.
Scientific research has begun to further elucidate the connections between the environment in which we live and our DNA. This branch of scientific interest is often referred to as epigenetics and has become an increasingly used and recognised term of biology. The term epigenetics itself is not new and was first published over 70 years ago by a developmental biologist Conrad Waddington. A paper published in 1942 in the journal Nature entitled ‘canalization of development and the inheritance of acquired characters’ Waddington sought to understand how “….genotypes of evolving organisms can respond to the environment in a more coordinated fashion”.
Epigenetics as a discipline has transformed how we think about genetics and our genome. It has been considered as the study of biological mechanisms which switch genes on or off. Unfortunately, the term epigenetics itself has been defined and used in different interchangeable ways leading to some ambiguity of its true meaning. The literal meaning of epigenetic is ‘in addition to genetics’ and refers to the molecular marking or ‘tagging’ of our DNA structures. Several biological mechanisms allow physical changes to the genome without altering the DNA code itself and are often referred to as epigenetic modifications.
Have you ever met identical twins – who despite being genetically the same – had different characters, behaviours or even physical traits?
So, what makes genetically identical twins different? M. Fraga et al. asked themselves the same question. Their explanation was as follows: “By using whole-genome and locus-specific approaches, we found that approximately one-third of MZ twins harboured epigenetic differences in DNA methylation and histone modification. These differential markers between twins are distributed throughout their genomes, affecting repeat DNA sequences and single-copy genes, and have an important impact on gene expression. We also established that these epigenetic markers were more distinct in MZ twins who were older, had different lifestyles, and had spent less of their lives together, underlining the significant role of environmental factors in translating a common genotype into a different phenotype.”

Looking at this picture of these two mice, would you think they are genetically the same?
The answer, surprisingly, is yes – they are, and this constitutes an example of the effect of food on epigenetics. All mammals have a gene called agouti. Individuals with the active agouti gene have yellow fur, tend to be obese and prone to diabetes and cancer. However, this gene can be deactivated by DNA methylation – an example of epigenetic modification. If pregnant yellow mice were fed a methyl-rich diet (a diet rich in nutrients such as folic acid, choline, betaine and vitamin B12), most of her pups were brown and stayed healthy for life. The offspring still carries the agouti gene but loses the agouti phenotype.
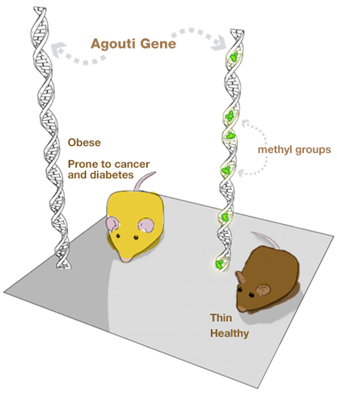
These results show that the environment in the uterus affects the health of adults and emphasize the role played by the mother’s diet in shaping the epigenome of her offspring. Summing up, our health depends not only on what we eat but equally important is what our parents ate.
How can we translate this knowledge into practice?
Imagine you did a genetic test. Its results indicate the presence of genetic variations, that contribute to reduced detoxification capacity and, consequently, to an increased risk of DNA damage. It is worth mentioning that the detoxification process consists of two stages:
– Phase I, leading to the formation of even more harmful metabolites than the initial toxins. For this reason phase I, should last as long as possible!
– and Phase II, as a result of which xenobiotics are transformed into a water-soluble form and can then be excreted from the body. Unlike phase I, this process should be as fast as possible!
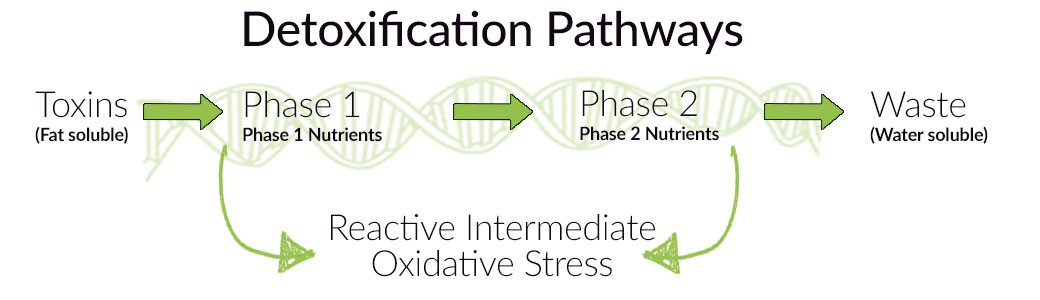
Fortunately, those variations included in your genetic testing, are actionable and there are lifestyle and dietary changes which can compensate. In the case of detoxification problems, we strive for a state of balance between both phases. So-called “load” of phase I should be reduced – in other words, the number of toxins should be decreased – and phase II should be supported. We can achieve this by:
- eliminating exposure to environmental carcinogens, such as smog, alcohol, cigarettes, smoked and grilled foods, pesticides and other contaminants.
- increasing the consumption of vegetables and fruits (organic, if possible). In this case, however, the most important is the daily consumption of 1-2 servings of cruciferous and alliums vegetables.
- including in the diet nutrients such as sulforaphane, resveratrol, curcumin, fish oil, quercetin, daidzein and lycopene. Several studies are showing that they support phase II detoxification
Learn more about nutritional genomics
Together, myDNAhealth and Qube Vision are offering Health and Nutritional Professionals a unique opportunity to join these pioneering online courses.
The Nutritional Genomics Interpretation programme comprises four courses covering the following topics:
- Molecular and Personalised Nutritional Genomics (foundation course)
- Carbohydrate Behaviours and Lipid Metabolism
- Immune Mediated Inflammatory Conditions and Nutrient Sensitivity
- Detoxification, Oestrogen Balance and Methylation
For more information and to buy your ticket, please visit the myDNA website: https://mydnahealth.co.uk/course-list/
Key Takeaways
- The correct state of nutrition provides our body with optimal conditions for everyday functioning and helps maintain our health
- Through scientific research it has become clearer that the response to a particular nutrient is not the same for each person. There are a variety of reasons for this such as metabolic, environmental and/or genetic.
- The knowledge we can apply from -omics technology, together with understanding of nutrient-gene interactions has developed a field known as Nutritional Genomics.
- At present, the nutritional genomic relationship is made up of two related but distinct fields namely, Nutrigenomics and Nutrigenetics. Nutrigenomics focuses on how diet affects gene expression. Nutrigenetics focuses on how the gene variants in our DNA influences our response to nutients i.e. how our genes determine the effects what our food has on us.
- The human genome was found to be approximately 3 billion nucleotides, around 22 000 genes, a large amount of information
- The literal meaning of epigenetic is ‘in addition to genetics’ and refers to the molecular marking or ‘tagging’ of our DNA structures. Several biological mechanisms allow physical changes to the genome without altering the DNA code itself and are often referred to as epigenetic modifications.
With many thanks to Dr Eve Pearce and Magdalena Puchalka from myDNA Health for this blog. If you have any questions regarding the health topics that have been raised, please don’t hesitate to get in touch with Amanda via telephone; 01684 310099 or e-mail amanda@cytoplan.co.uk
About the authors of this blog
Dr Eve Pearce PhD (medicine), DipION, mBANT, CNHC
Before joining myDNAhealth in 2016 as Scientific Officer, Eve worked extensively in the field of Human Genetics and has published papers in both cardiovascular and cancer research. A passion for nutrition and wellness led her to further train as a Nutritional Therapist with the Institute for Optimum Nutrition.
Eve is also a module leader at the Institute for Optimum Nutrition and British College of Osteopathic Medicine, specialising in delivering nutritional education with a Functional Medicine approach.
Magdalena Puchalka, MSc Nutrigenomics, Registered Clinical Dietitian
Magda joined myDNAhealth in 2019 and is experienced in dietetics, nutrigenetics and completed an internship in a genetics laboratory.
Last updated on 23rd February 2021 by cytoffice



What is the nutritional genomics course? Is it available?
Nice article. Perhaps a mention of Bruce Aimes work and landmark paper in the Journal for Clinical Nutrition in 2000, on this subject would have been relevant. Genetic faults can be compensated for by supplementation. That is the good news.
Hi Steve,
Details on the course and a link for more information can be found at the bottom of the article. We’re pleased you enjoyed the blog post, thank you for your comment.
Thanks,
Jo
I saw the title and thought oh maybe they are offering a DNA test to identify best nutrient supplements, that would be useful. I just couldnt read though so much information. For me it was not written in an interesting way so I stopped reading
Hi Barbara,
Thank you for your comment on our blog. Some of the articles we feature on the blog can be more technical than others, as we try to strike a balance with the content to suit a wide and varied audience. We will take your thoughts into consideration.
Many thanks,
Jo
I enjoyed reading this as a biomedical genetics research scientist between 1992 and 2014 involved in the human genome mapping effort and now a holistic therapist, sharing these principles to my clients; I had the great pleasure of meeting the lead author of the landmark Nature paper on Agouti mice at the time of publication which truly shaped my future professional direction.
Thank you for this very clear and interesting introduction to nutritional genomics – sometimes we just need the very basics to start to take our knowledge forward!
Excellent article overall and I’m seriously thinking of doing the course. However, I have a query about Phase I and Phase II detoxification. I believe I understand the concept, but I would have thought that slowing Phase I is better than having it continue as long as possible (which would allow the more harmful metabolites to accumulate – surely that is not what is wanted?). I think I know what was meant, but the way this has been put is misleading. Furthermore, saying ‘Phase II, as a result of which xenobiotics are transformed into a water-soluble form…’ indicates that it is in Phase II that biotransformation of the lipophilic toxins into more water-soluble compounds occurs. That is not what I was taught at university, nor is this in agreement with the Functional Medicine Textbook Chapter 31 (2005, 2006). The biotransformation occurs in Phase 1 via the Cytochrome P450 enzymes, doesn’t it?
Hi Isabella,
I do concur that it is rarely beneficial to upregulate phase I, as that can result in a greater level of harmful toxic metabolites being produced, particularly if there is poor synchronicity between phase I and II enzymes. Phase I, II (and III) reactions may occur simultaneously or sequentially. The biotransformation of lipophilic toxins into more water-soluble compounds occurs in phase I but this can be simultaneous with phase II.
Thanks,
Amanda